
Audio file
(DIRECT CURRENT THEME PLAYS) (FADE IN MELLOW ACOUSTIC GUITAR MUSIC) MATT DOZIER: Hey everyone, welcome to Direct Current, I'm Matt Dozier. CORTNEY KREER: And I'm Cort Kreer. DOZIER: We've been hard at work on new episodes of the podcast, and we've got a fun one for you today. KREER: So Matt, you went to a big conference in DC to talk to some scientists recently. DOZIER: Yep — AAAS. That's the American Association for the Advancement of Science, and they have this huge annual meeting where scientists from all over the world get together to share their findings, discuss research techniques, talk about how to communicate about science, that sort of thing. KREER: Yeah! And there's tons of folks there from the Department of Energy and our 17 National Labs. DOZIER: Absolutely. And so I got a chance to sit down with a couple of them to talk about a subject I find really fascinating: microbiomes. KREER: Microbiomes! So like.... biomes, but super small?! Kinda? DOZIER: Kinda! So, there's this invisible world of microbes that live all around us, on our skin, in our guts, and even in the ground under our feet -- and we're only just beginning to understand their function. The researchers I interviewed are especially interested in using genetic data to study the microbiome of soil. Turns out, these tiny organisms can help us answer some really big questions. KREER: Sounds awesome! After the break, we'll listen in on that full conversation, recorded live on stage at AAAS 2019. Stick around! (MUSIC FADES OUT) MATT DOZIER: Hello and welcome everyone, this is Direct Current - An Energy.gov Podcast. My name's Matt Dozier. We're here at the AAAS Annual Meeting at the Sci-Mic Podcasting Stage. I'm with the U.S. Department of Energy, and I'm joined here by two scientists. From the Joint Genome Institute, we have Dr. Susannah Tringe, and Dr. Jason McDermott from Pacific Northwest National Laboratory. Thank you both for joining me today. SUSANNAH TRINGE: Yeah, thanks for having us. DOZIER: I would like to have you first just give us a little introduction, tell us about yourselves and your research. Susannah, want to start with you? TRINGE: Yeah, sure. I work at a place called the Joint Genome Institute, which is a user facility that does genomics work for the user community, and my own area of expertise is called microbial community genomics using DNA and RNA sequencing to study whole communities of microbes. And I particularly focus on wetland soils and agricultural soils. JASON MCDERMOTT: Yeah, and I'm from Pacific Northwest National Laboratory, which is in Richland, Washington. A lot of people confuse that with being in Seattle, but we're actually on the east side of the state. I'm a computational biologist — I'm a microbiologist by training, and I've kind of gotten back into microbiology through working on soil microbiome projects more recently. DOZIER: Well, I appreciate you both joining me here today. We are here to talk about microbiomes, and I think just for basic background, could you help me understand what a microbiome is? TRINGE: Yeah, sure. So, microbes are found in all kind of habitats, ranging from our own bodies in our guts and on our skin to environments like soil or water, and usually there's a whole spectrum of organisms — not just one microbe, but a whole community that works together. So the microbiome of soil, or your gut, is sort of a community that's thought of as a unit that works together. DOZIER: There is a lot of talk, a lot of attention paid these days to the human microbiome, as you said — with gut microbes, that sort of thing. Now, your work specifically is more focused on soil, right? TRINGE: Right. DOZIER: So why study the microbiome of soil? Isn't dirt just dirt? TRINGE: So there's actually a lot of important processes that take place in soil that relate to carbon and nutrient cycles, supporting plant growth or the animals that live there. So we're really interested from the energy perspective in how soil is involved in the production and uptake of greenhouse gases. DOZIER: Right. OK, so Jason, I want to get to your research in a bit, but first, when it comes to the actual collecting samples and figuring out what is in the soil microbiome, how do you actually go and collect those samples, Susannah? TRINGE: Yeah, well, in the wetland projects, we have a corer. So we can go out and take these cores of soil, usually about a foot or two deep. And then we take that and usually mix it up and freeze it to take it back to the lab, where we can then extract DNA for sequencing or RNA. Another thing we do with the cores before we extract the nucleic acids is actually measure their ability to produce and uptake greenhouse gas. We have an instrument we can put the core in and measure its production in real time so that we can really see that that piece of soil is carrying out this activity, and then do our molecular assays to see how that matches up. DOZIER: So you're not just using basic gardening tools, right, to collect these samples? TRINGE: Yeah. Yeah. You have specialized corers, and you use lab equipment when you want to keep it clean, and package it and put it on dry ice and get it back to your lab. DOZIER: You want to keep the dirt clean? TRINGE: As best you can. Especially you don't want any of your human-associated microbes getting mixed in with your soil, and they're all over you. So you do want to prevent that. DOZIER: Right. So it's actually pretty clean work when it comes to collecting and transporting the samples. TRINGE: Yeah. DOZIER: So, tell us what you do with these samples once you get them back to the lab. What is the goal here, and how do you do that work? TRINGE: Yeah. So the soil samples, in order to extract DNA, there's a series of chemical steps that works amazingly well with a variety of samples. The protocols have been worked out such that you can take it, you usually add some beads to break all the cells up, and you vortex it, which is kind of shake it really hard, and then put it through a set of purification steps that take all that soil and dirt away, and take out all the proteins. If you're looking for DNA, get rid of the RNA as well, and finally end up with pure DNA that you can use for sequencing. DOZIER: Gotcha. And Jason, maybe you can jump in here, too. So this data that you've collected once you've sequenced all of the genes in this sample of whatever it is — soil or something else — what does that look like to you, in terms of the sheer quantity of data? MCDERMOTT: It's a pretty overwhelming amount of data. If you think about what you might open up in an Excel spreadsheet or something like that, you can think of that multiplied many, many times over. And it's something that you can't really deal with with the kinds of normal bioinformatic tools that exist. So you have to use in some cases high-performance computing — that's like big compute clusters — to work through some of the data. Especially from samples like soil, which are very complex, you end up having just a lot of data there. I should also note on Susannah, she said they take out the proteins to discard them. At PNNL, that's one of the things that we focus on, is taking those proteins and some other kinds of small molecules and also characterizing those to kind of layer on top of the metagenome information that's coming out. DOZIER: Right. And I think it's important to define that term, the metagenome. So there's the genome — you're talking multiple genomes, right? I think people might be familiar with the sort of personal at-home genome tests. Is this anything like that? TRINGE: Yeah, yeah. The basic technologies for sequencing are similar regardless of the application, and although many of the technological advancements have been focused on producing cheaper human genomes, the whole rest of the field of genomics has benefited from those changes in technology. In terms of a metagenome, that is sort of a coined term using that prefix "meta" in the same way you might talk about a meta-analysis being a combined analysis of many datasets, the metagenome is the combined genome of many different organisms present in a sample. DOZIER: Right. So once you have all that data, you have the massive super-spreadsheet of all the fragments of genetic data, how do you go about making sense of all of that information? Whoever wants to take that, I'm sure there's a lot. MCDERMOTT: I mean it's a really difficult problem, and one of the problems is that if you look at samples like human samples or something, the genes of humans are fairly well-characterized. We pretty much know what the functions of most of them are. With a metagenome from a microbiome, we have lots and lots of protein-coding sequences, so these are the bits of genetic code that actually code for things that do work, and we really don't have a good handle — I mean, some of the estimates are as high as 80 percent, 70 or 80 percent of the pieces, we just don't know what they do. And a lot of them are not similar to anything else that we've seen. And so it's a difficult process. So the first pass is probably just to match things up with things that have already been seen. So you can kind of go through and say oh, this is definitely a certain kind of bacteria that we've seen before. You know something about what the bacteria does, and then you kind of go down the line of, like, "OK, here's a bunch of stuff that we know what the function is, but we can't really attribute it to any particular organism. It comes from a particular bacteria, but we don't know where that comes from. And then there's just this whole bunch of dark matter, which is relatively unknown. We can look at it with the current tools, and we just don't know what it is. DOZIER: And you're using dark matter kind of as a euphemism there, right? MCDERMOTT: It is definitely a euphemism. So what I was using it as is basically just the unknown genetic potential of microbiomes. DOZIER: So Susannah, specifically in soil, we were talking earlier that basically we don't fully understand what soil is made up of. Is that generally correct? TRINGE: Yeah. A lot of the organisms there are organisms that we are not able to cultivate in the lab, and the way we've learned that they are there is through these molecular techniques. We can see the same sequences showing up in our DNA sequencing, or at least the same families, and we can look at particular genes that help us place it on a sort of phylogenetic context, but ultimately they might not be at all closely related to the things that we've grown and sequenced, and know what they do. DOZIER: In say, a typical sample of soil, of all that information that you collect and get, do you — how much of that can we actually interpret at this point? TRINGE: Something like less than half. Even if you can — depending on what you consider "interpret." You'll take your data, you'll clean it up, you will try to assemble it into genomes if you can — which for a single genome would always be the objective — to get a good quality assembled genome so you know not just each individual read but how they go together. And then annotate genes, these protein-coding sequences on it. And we can... for the metagenomes, it's not that hard to say, "this is probably a protein" because it could code for an amino acid sequence, often you can say it's a protein, and it's similar to something else that's been sequenced some time, but the fraction that you can actually say, "It's similar to a protein that someone has tested in the lab and carries out this function," is maybe 40 percent or something of the proteins that you see there. DOZIER: Right. And you said that a lot of these microbes in the soil, we can't cultivate them in the lab, right? So we then can't really study them thoroughly. Is that basically right? TRINGE: Yeah, exactly. What you'd love to do is test your hypotheses, say, "I think this organism can eat this particular carbon substrate." What you'd like to do is grow it in the lab and feed it that carbon substrate and see that it takes it up and does something with it, but in actual practice, since you can't even get it by itself, you don't have any way of testing that kind of hypothesis in a simple, straightforward way. DOZIER: Right. OK, so with all of these unknowns, with all of this data, what are some of the challenges and some of the ways you try and tackle sorting through, comparing all this data, and figuring out what it means? TRINGE: Well, one of the most useful things that comes in is really having a great reference database. Because like we said, the first thing you do is compare it to everything else you have and try to say are there other things that are close enough that I can kind of jump that gap and say, I think it carries out this function. And so, having as many genomes, just isolated microorganisms as possible, really helps us interpret it. But also, being able to compare the whole spectrum of genes that you have present in your sample with what people have seen in other samples that are hotter or colder, or wetter or dryer, can really help you sort out — there's thousands and thousands of genes, which ones might be the important ones in your particular system? So the comparison is really the most powerful tool we have for these data that we have a hard time interpreting. DOZIER: Right. And Jason, that's something you're involved in, right with the comparative analysis across these databases and that sort of thing, is that right? MCDERMOTT: Yeah. That's true, I mean we're working on ways to extend some of the existing methods for looking at similarities. So there are very good ways of identifying similar sequences, so these are the gene sequences or the protein sequences to tell you what this unknown protein is similar to. But like I said, there's still a lot of territory, and there are a number of cases where we know that the proteins have the same function, but we know that their sequences are very different. And so how do you kind of try to take those existing methods and extend them so we can get more insight into what is actually going on. DOZIER: With so many unknowns, does it every feel frustrating or overwhelming to just look at all this data and say, "What does it all mean?" MCDERMOTT: I think it's really exciting, actually. I mean yes, there's an element of frustration in that you'd like to push further, and sometimes you have things that you're like, "Oh, I really want to know more about this system." But it's really interesting to think about the potential of this data and the things that we don't know. We're working at PNNL on a project where we're trying to characterize some of the viral sequences that are in this. So viruses actually infect bacteria. There's a whole class of viruses that infect bacteria, and we can identify those from those metagenomes from the microbiomes, but we really have, like, zero idea... very little idea of what's going on with those, why those viruses are there, and what they're doing in terms of the dynamics of the microbial communities. DOZIER: So that's even beyond the microbiome, that's the virome, right? MCDERMOTT: The virome, yeah. DOZIER: You touched on things we can do with this, and exciting opportunities. Susannah, tell me a little bit about what you're trying to learn from this research. What are some of the big things that you're able to shine more light on? TRINGE: Yeah, there's a lot of different things we're trying to learn, but one of the main motivations behind my wetlands work is not just better understanding the carbon cycle, but identifying ways that we can help shift the greenhouse gas balance in the atmosphere. Wetlands in particular are known to be very effective carbon sinks, and they have vegetation that takes up a lot of carbon dioxide and converts it into plant matter, but they can also be greenhouse gas sources. They produce methane at times, or just CO2 from breaking down the plant matter, and that's a totally microbial process. And so there's potential for it to be a carbon source or sink. When that is the case is not that well understood. There's sort of general trends, but any given wetland is hard to know where it's going to fall on this spectrum. And there's a lot of wetland restoration going on now, usually aimed at improving habitat, or water quality, or flood control — not usually toward carbon sequestration. I think all of those wetland restoration projects have the potential to be carbon sinks, but only if we really know how we can make them carbon sinks and not carbon sources. So we're trying to look at the microbes and say, well this sample produced a lot of methane, this one's not producing much at all — what are the biogeochemical and genetic factors that we can correlate with that pathway, and then can we step back to what was the environment that made that conducive to that particular balance? DOZIER: So for your wetlands work, I think some folks might be familiar with the general area if they know the Bay Area at all. TRINGE: Mmhmm. DOZIER: Tell us a little about where you've been doing that research. TRINGE: Yeah, so we're located in the Bay Area, so it was a perfect place to study wetlands in this bay-delta region. Our initial studies were up in the Sacramento-San Joaquin Delta, where there used to be a whole range of wetlands, but almost all of it has been converted to other uses, mostly farmland, because they have very rich organic soils that make great farming land. Unfortunately, that wasn't a very sustainable process because that organic soil tends to break down once you've drained it. It's metabolized into carbon dioxide, it gets compacted by the farming equipment, so if you go out to these islands it's very striking that they're very far below the surrounding water and prone to flooding, because if the levees breach they become flooded. So the wetlands we started studying there were partly to try to build back that subsided land. We've also been, in terms of studying this whole question of carbon cycling, we've been sampling wetlands throughout the Sacramento Delta and the San Francisco Bay region, and most recently the salt ponds in the San Francisco Bay, many of which are slated for restoration, so it gave us an ideal opportunity to compare unrestored and restored wetlands. DOZIER: And I think I've seen those from the airplane flying into SFO. TRINGE: Yeah. They're very striking. They're very bright red because they have these halophilic microorganisms that produce red pigments. DOZIER: Moving on from that, what is for each of you the most exciting thing about your research that you're working on right now? TRINGE: I think for me, it's really the potential to have an impact on our understanding of the environment and our stewardship of the environment, being able to improve the way we manage our ecosystems. But also, just this incredible power for discovery, that you sequence almost anything out in the environment and there's still a whole bunch of it that's not understood, and that can be frustrating, but you also think, imagine if we really did know what all of this was. There are probably some amazing discoveries just lying in wait if you can figure out how to interpret the data. MCDERMOTT: Yeah. And that's really exciting for me, as well, as I expressed. I think one of the key parts of that that we're really interested in — and I know JGI is interested in, as well — is the open science idea, which is trying to make as much of this data available and usable by the scientific community. Because we have specific questions that we're asking about it, but if we can put it out there and have other researchers come and ask different questions, they're going to get really interesting discoveries that we never even thought of. So part of that is moving that data forward and putting it out so that we can enhance the biological discovery. I'm really excited about how we take different kinds of data, so the metagenome data but also as I mentioned we have protein measurements that are kind of similar, and small molecule measurements, and how we knit those back together into a picture of an entire microbial community. And you can kind of think of it like this is a very interdisciplinary approach, like your modern medicine, where you go in and you see a doctor, but you're not just seeing that general practitioner. You're seeing an X-ray technician, you're seeing a nurse, phlebotomist, you're seeing a bunch of people with a bunch of different expertise and how do you take each of those pieces that you get from that and knit it back into a diagnosis of your problem? That might be, how do you keep carbon in a form that's not in the atmosphere, or things like, how do we enhance agricultural production, which is something we're interested in from our soil work. DOZIER: Sure, or biofuel crops, that sort of thing. MCDERMOTT: Biofuel, yeah. DOZIER: You mentioned briefly the importance of partnerships. Obviously there's the interdisciplinary work, but also the open science, bringing other researchers, other institutions in to kind of help tackle this enormous challenge in interpreting all this data. Tell me a little about that and the way that DOE and the National Labs have been working to faciliate this work across disparate sites and institutions. TRINGE: Yeah. Well, like you said, some of the data can be overwhelming, and that means when you are doing your analysis, you have to really go after a question, but then someone else may have a completely different question that will be answered by your data that they will approach in a totally different way. Some of the incidental discoveries have been really amazing, like these "CRISPR" pathways were first discovered in microbial genomes. That was using data from sequenced genomes, and no one had to generate new data in order to discover that pathway. So we've tried to always make our genomic data public, and I think now with all these other types of data, similarly, you might integrate them in different ways. You have these metaproteomics, or metatranscriptomics, and metagenomics, and having all of those available in one place where you can pull them in and come up with creative strategies for analyzing them is really powerful. So there's an initiative by DOE to develop environments where you can do better analyses, as well as databases that store all of these omics data and allow you to retrieve them and compare them in the ways that you want to. MCDERMOTT: This takes a lot of coordination across sites with different expertise. We're certainly partnering on our soil project with JGI to look at the virome, as I mentioned. But in some other cases, as well, just using the data and resources that they offer. TRINGE: Yeah, and it's also worth nothing that both EMSL and JGI are user facilities, and so we're not just doing the sequencing or characterizing or looking at the samples that we find interesting. We're putting out calls to the user community to come use our capabilities. JGI has a program called the community science program that's just open to anyone doing research in energy and the environment in the DOE mission spaces, and they can come to us and work with us to use our capabilities, and the same with EMSL. MCDERMOTT: Same with — yeah. EMSL is the Environmental... uh... TRINGE: Molecular... MCDERMOTT: (LAUGHS) Molecular Sciences Laboratory. Sorry. And it's colocated at Pacific Northwest National Lab. So it's very similar to JGI. DOZIER: And I'm sure there are folks here at AAAS this weekend that have been involved with both of your institutions, as well. TRINGE: Yeah. Yeah. Definitely. And all of those data from those projects are made available to the scientific research public in this spirit of open science, so that those discoveries can really be leveraged upon each other. MCDERMOTT: And I think it's really important to note that those kinds of efforts have really been driving forward the field. We're closely associated with EMSL, but we're not in EMSL, but because we're using those resources, all the data that we're making has to be publicly available, has to be open, has to be reusable. And so that's been a really great thing to be working with. DOZIER: Well, thank you to you both. Susannah Tringe, Jason McDermott, really appreciate you joining me today. TRINGE: Alright, thank you. MCDERMOTT: Thanks very much. DOZIER: Thank you to AAAS for having us here on the Sci-Mic podcasting stage. This has been Direct Current - An Energy.gov Podcast, I've been your host, Matt Dozier. You can find us at energy.gov/podcast or wherever you get your podcasts. Thank you all for listening! (APPLAUSE) (FADE IN SOFT ELECTRONIC OUTRO MUSIC) DOZIER: Thanks again to AAAS for having us. Thank you to the folks at the Joint Genome Institute, Pacific Northwest National Laboratory, and the Department of Energy's Office of Science for helping arrange this interview; to Kirsten Hofmockel at PNNL, who was supposed to join us but had her flight from Washington canceled at the last minute; and, of course, to Susannah Tringe and Jason McDermott. KREER: As always, if you've got a question or want to leave us some feedback, email us at directcurrent@hq.doe.gov, or tweet @energy. And if you're enjoying the show, share it with a friend and leave us a review on iTunes. We read them, and we listen. KREER: Direct Current is produced by Matt Dozier, Paul Lester, and me, Cort Kreer. I also create original artwork for every episode, which you can find on our website. DOZIER: Additional support from AnneMarie Horowitz, Ernie Ambrose, Gigi Frias, and Atiq Warraich. We’re a production of the U.S. Department of Energy and published from our nation’s capitol in Washington, D.C. KREER: Thanks for listening! (MUSIC FADES OUT)

Recording live at the 2019 AAAS Meeting in Washington, D.C., on Feb. 17, 2019. Left to right: Matt Dozier, U.S. Department of Energy; Susannah Tringe, Joint Genome Institute; Jason McDermott, Pacific Northwest National Laboratory.
There's an invisible world of microbes living all around us — on our skin, in our guts, even in the ground under our feet.
We're only just beginning to understand their function. In this episode of Direct Current, recorded live at the 2019 American Association for Advancement of Sciences (AAAS) Meeting in D.C., we interview two scientists who are using genetic data to study the "microbiome" of soil. Turns out, these tiny organisms can help us answer big questions about agriculture, climate, and much more.
Marsh Madness
DOE Joint Genome Institute Metagenome Program Head Susannah Tringe and postdoc Susie Theroux discuss their research on microbial diversity of marshes in the San Francisco Bay Area.
Video courtesy of the Joint Genome Institute
Dr. Susannah Tringe is the Deputy of User Programs at the Joint Genome Institute (JGI), a Department of Energy Office of Science user facility managed by Berkeley Lab. Scientists from around the world come to JGI to work on genomic research advancing fields like clean energy and environmental restoration. Watch the video for a great breakdown of Susannah's own research on tidal marshes in the San Francisco Bay Area.
Drawing Conclusions
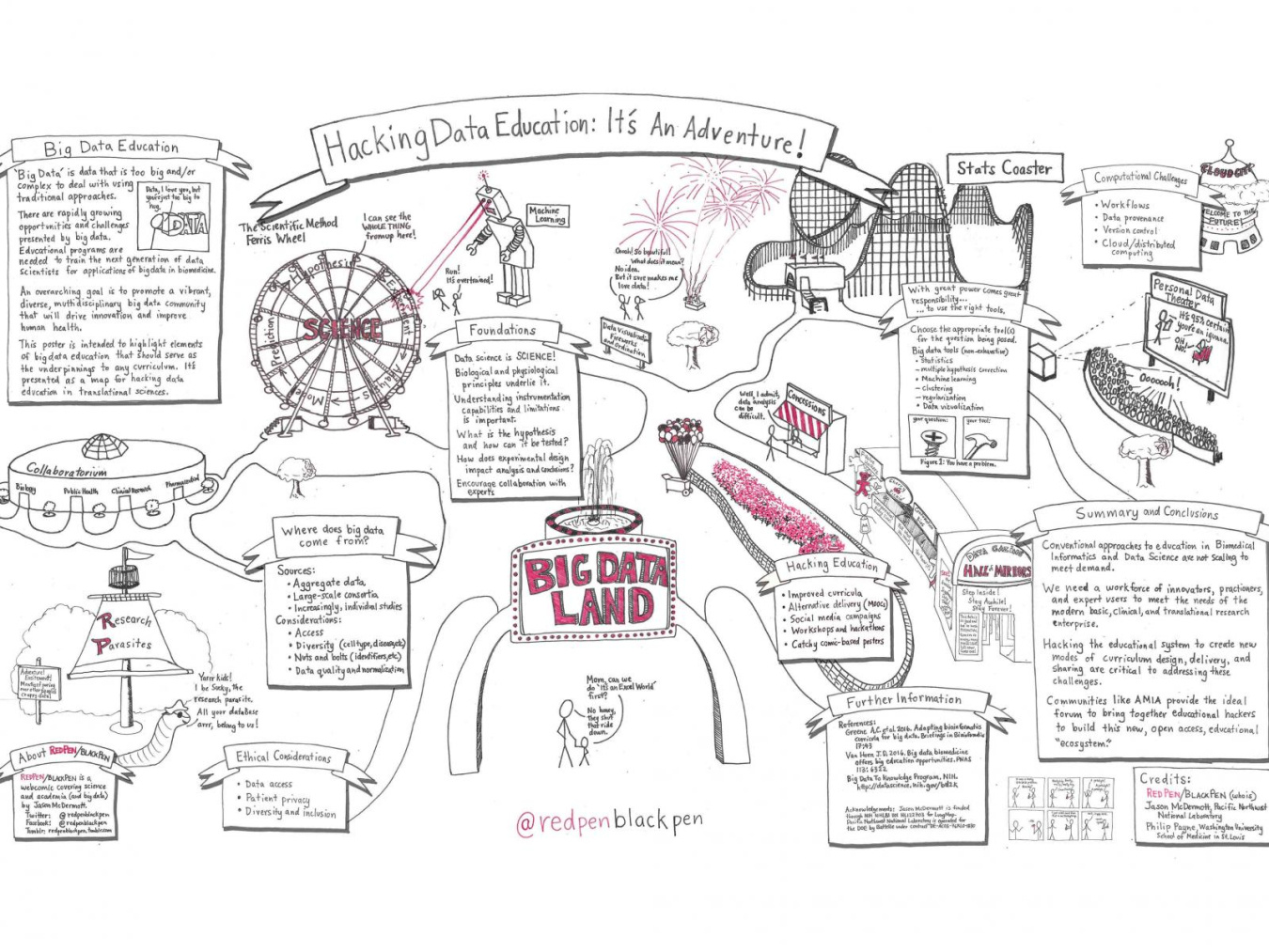
Our other guest, Dr. Jason McDermott, is a senior research scientist at Pacific Northwest National Laboratory, where he uses computer modeling and systems analysis to tackle tough questions in biology (like what's in the soil microbiome!). When he's not busy crunching massive amounts of data, he's got a second passion: using comics and art to help communicate science! Learn more about Jason's double life as a scientist and cartoonist.
Unlocking the secrets of microbes is just one way Energy Department and National Lab scientists use genomic science to solve some the world's must urgent energy and environmental challenges.